Innovative method for detecting co-occurring histone modifications in a single step using hybrid proteins
Abstract
Due to the role of epigenetics in a vast array of diseases, the study of epigenetic mechanisms has become one of the most rapidly growing fields of biology.
Amongst others, epigenetic alterations involve post-transcriptional histone modifications (PTMs). Occurring in complex patterns, they form the so-called ‘histone code’. In order to decipher said code and unravel its involvement in disease, the characterization of co-occurring histone modifications is considered to be an important cornerstone.
An innovative method now allows the detection of co-occurring histone modifications in one single step. Contrary to conventional methods, the technique offers much improved sensitivity, is easy to perform and features consistent quality.
Background
The last two decades have seen an enormous increase of insight into the role of epigenetics in a vast array of diseases such as cancer, mental retardation and diabetes. In fact, the study of epigenetic mechanisms has become one of the most rapidly growing fields of biology, generating growing interest of the pharmaceutical industry.
Amongst others, epigenetic alterations involve post-transcriptional histone modifications (PTMs). Occurring in complex patterns, they form the so-called ‘histone code’. In order to decipher said code and unravel its involvement in disease, the characterization of co-occurring histone modifications is considered to be an important cornerstone. The invention at hand offers an innovative approach taking their analysis to another level.
Problem
Currently, co-occurring histone modifications can only be studied by the isolation of nucleosomes using consecutive chromatin immunoprecipitation (ChIP). Utilizing antibodies, the modifications are subjected to several isolation steps.
This technique carries several serious drawbacks. It is time consuming, requires a lot of starting material and is difficult to perform. Furthermore, ChIP assays have poor sensitivity and, at present, DNA gained from the process cannot be investigated using next-generation sequencing.
Solution
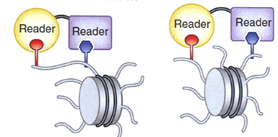
Scientists at the University of Stuttgart developed an innovative method, in which two or more highly specific histone modification binding proteins are fused to form bi- or multispecific hybrid proteins. This construct allows the detection of co-occurring histone modifications (either on the same or different tails of one nucleosome) in one single step (see Figure). The DNA gathered from the process can be analyzed using next-generation sequencing. Thus, it enables the investigation of co-occurring histone modification patterns on a genome-wide or locus-specific scale.
Contrary to conventional methods, the technique offers much improved sensitivity, is easy to perform and features consistent quality. Moreover, it requires only modest amounts of starting material.
Advantages
- Detection of co-occurring PTMs in one single step
- Contrary to conventional techniques consistent quality
- Technically easy to perform
- Less time-consuming and less starting material needed than in conventional antibody mediated techniques
- No animal testing necessary in the production process
- Can be analyzed using next-generation sequencing
Application
Detection of co-occurring histone modifications.